The second Virtual Research Environments (VRE) Workshop was held by Zoom on the morning of the 6th May and featured two relatively new areas of archaeology – Ancient DNA and Environmental Data & Research. After a welcome by Franco Niccolucci (Project Co-ordinator), the workshop host, Guntram Geser (Project Innovation Manager) set the scene by summarising the results from the first VRE workshop. One new outcome since this was held is an upcoming meeting with the THANADOS Project to discuss possible collaboration.
Guntram also presented a snapshot of recent developments in ancient DNA (aDNA) studies, which is a rapidly expanding field of research, as illustrated with figures of searches of DataCite and OpenAIRE. There is an expansion from the initial focus on remains of hominids and large-bodied animals to various animals and plants, bacteria, and sediments. In recent years, in addition to newly excavated remains, ever more samples for aDNA studies are sourced from various natural and cultural history collections.
Understanding the complexity of aDNA studies and the importance of contextual information
The format of the workshop was two presentations on each topic followed by discussion and the first was an overview of the study of ancient DNA (aDNA) provided by Eugenia Tabakaki from IMBB-FORTH. Eugenia highlighted the many uses and also issues faced by researchers in the fast-growing field of archaeogenetics. In a fascinating presentation, she explained how aDNA is used not just from humans and animals but also plants and bacteria to study several scientific questions that were impossible to answer with conventional archaeology methods. Eugenia also pointed out the dependence of sample preservation and genetic analysis with the natural environment. Studies include looking at humans, both as individuals and as societies, plants and animals and the extinction or expansion and migration of all of these, along with the impact of changes in cultural practices, infectious diseases, etc.
Large amounts of data are generated before the sample reaches the lab for aDNA analysis
This is a highly multi-disciplinary area involving experts from biology, geology, chemistry, to name just a few, and which generates large amounts of data even before the samples reach the lab. Challenges such as degradation and contamination of samples are commonplace and the context of biological material is also very important to be able to get meaningful research results. An important goal for aDNA researchers is an open environment for bio-archaeological data which is inter-linked, discoverable and accessible.
Maria Theodoridou, from ICS-FORTH, then presented the aDNA Application Profile which aims to support linking of the sample data with contextual and other information. A typical application would be to examine bone and teeth samples from an ancient necropolis using aDNA techniques to determine genetic sex, genetic diseases (e.g. sickle cell anaemia and thalassemia) and pathogens resulting from malaria. The aDNA workflow is intensive and starts with sample selection followed by DNA extraction, itself a complex process. Once this has been completed, the next step is to build and prepare libraries (collections of DNA fragments that have been treated with specific adapters, in a way that they are discoverable by sequencers). This then enables quantification through real-time PCR (a method for amplification of specific DNA molecules in real-time) and finally indexing and amplification using PCR (Polymerase Chain Reaction) followed by the actual sequencing of the ancient genome. Having generated a very large quantity of scientific data, specific bioinformatics tools are applied, depending on the scientific question, and the researcher can finalise their analysis and publish their results.
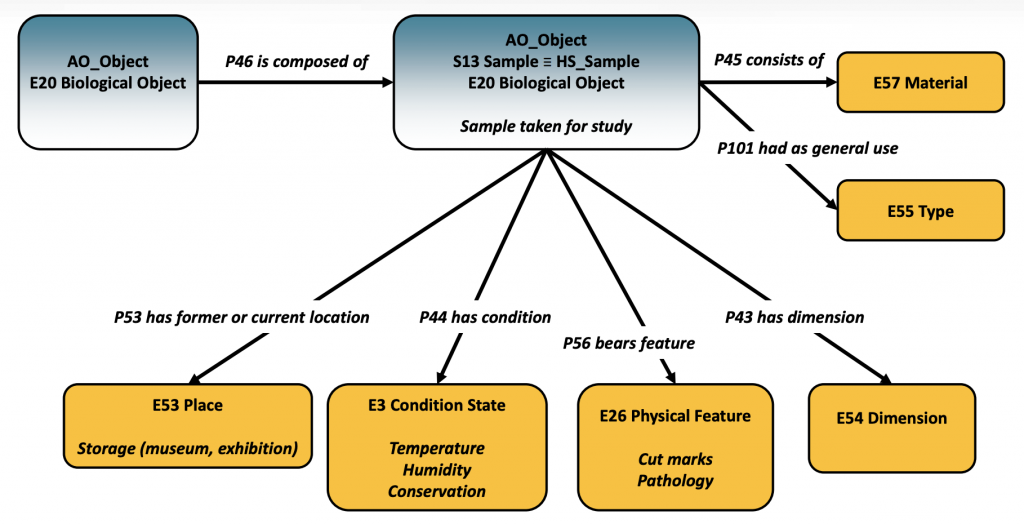
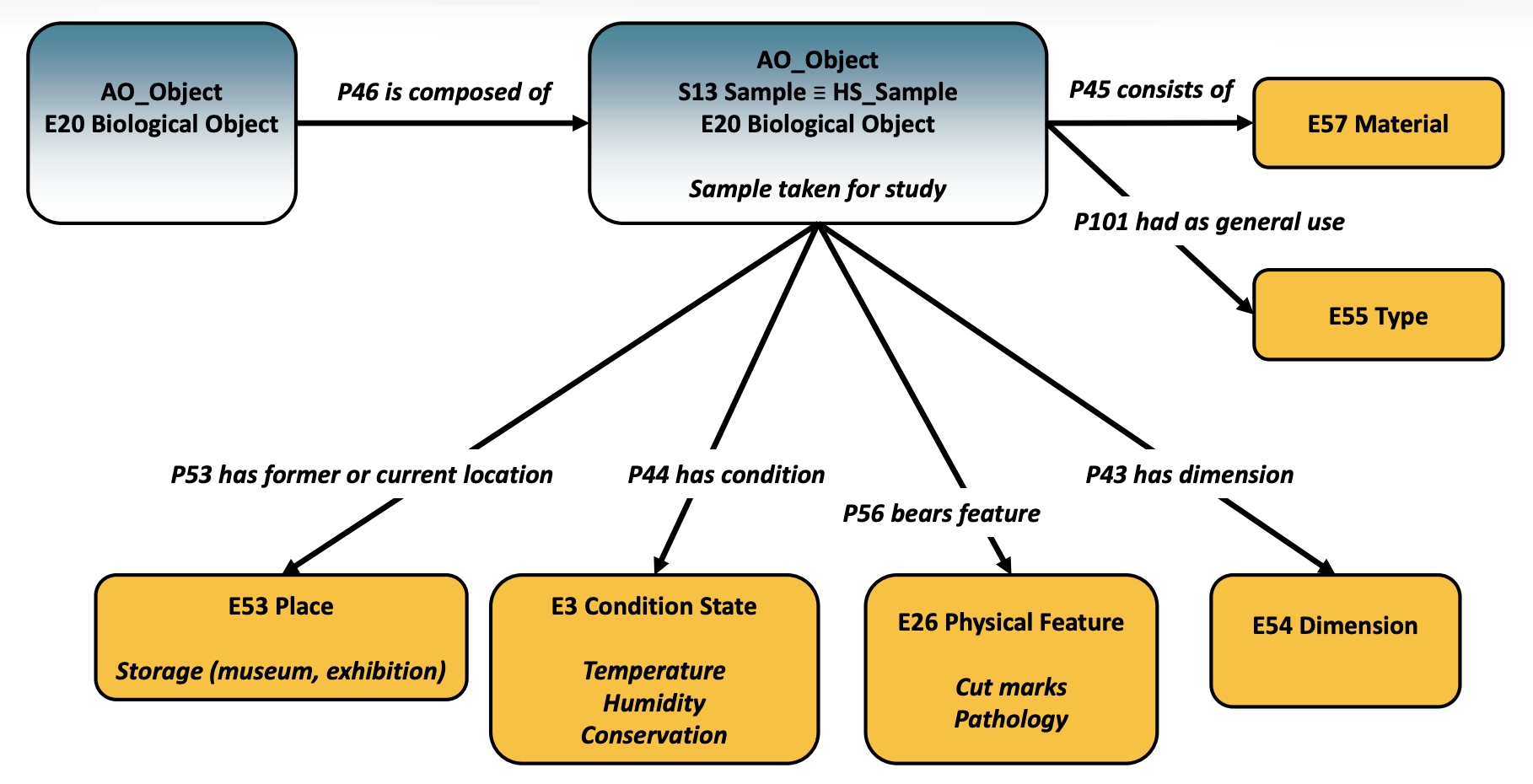
aDNA Application Profile mapping for a sample
Regarding the aDNA Application Profile, this is based on CIDOC CRM and a number of compatible models widely used in archaeology and is aligned to the ARIADNEplus Catalogue Ontology (AO-Cat) and CRM for heritage sciences (CRMhs). Maria then presented the modelling of a sample that will be used for aDNA analysis, using the classes S13 Sample and E20 Biological and the relevant properties for recording all the contextual data such as the details about the archaeological excavation.
She also briefly addressed the metadata model of the AncientMetagenomeDir, a community curated resource of lists of all published shotgun-sequenced ancient metagenome or microbial genome-level enriched samples. The metadata model could be mapped to the AO-CAT to include their data records in the ARIADNEplus catalogue and portal.
The discussion following the aDNA presentations started with a question on the potential to use the CRM class E92 Spacetime Volume to document how samples move from the fieldwork or a museum collection on to become derived sources and analysed results of aDNA studies. It would have to be seen whether this could work in practice, using an extended aDNA Application Profile, with Maria stating a preference for following general ARIADNEplus principles of keeping things simple.
The point was raised that in order to produce reliable, validated results using aDNA analysis, it is important to record as much contextual information as possible together with all the information about the processes during the analysis , e.g. for evaluating good or bad pre-treatments. As well, to be able to retrace analytical steps for repeatability and also to be able to decide which interim data to preserve. Contextual information helps unlock insights and can lead to more informed and accurate decisions on the research level. If researchers have access to valid, accurate, consistent, complete & reliable contextual data they can build a culture of integrity in the field of aDNA: The integrity of aDNA research question, sampling and interpretation.
Environmental reconstruction and long-term human ecodynamics
Following a short break, the workshop resumed with Philip Buckland and a presentation on Environmental Reconstruction and SEAD, the Swedish Environmental Archaeology Database. This involves aggregation of data through to analysis with links to other projects and systems, and both raw and analysed data in order to reconstruct landscapes at specific times in the past based on a very wide range of data, e.g. climate, geological, biological etc.
The lab and analysis data flow have to be described and also stored for which a method description is used. An example using ecological and species distribution data for beetles was used where the interpretation was needed for archaeology rather than agriculture and forestry. The database allows different levels of linking, e.g. by the same species of beetle, by its habitats… which is helpful to overcoming barriers such as agricultural classifications and dates are important too as environments change over time. Aggregated data of different databases are used for climate reconstruction and creating maps.
Phil also pointed to the initiative of the EarthLife Consortium to make all paleobiological data easily discoverable, accessible, and analysable. Currently this includes the Neotoma Paleoecology Database, the Paleobiology Database and the Strategic Environmental Archaeology Database (SEAD). The EarthLife Consortium promotes the sharing and use of paleobiological data through bridging databases with a common Application Programming Interface (API) and shared vocabulary.
The following discussion was on the technical feasibility of providing tools for an Environmental Archaeology VRE. Pasquale Pagano (CNR-ISTI InfraScience) suggested that working environment for using R scripts with Jupyter notebooks could be provided along with ARIADNEplus services and computational facilities required to process the data. However, it should be noted that the technical knowledge and know-how of the average user did not include R or Python skills and most lacked of time to acquire them plus lots of training materials are required. The point was made that this sort of training should be provided by Universities etc. (not ARIADNEplus) but if the project can show what can be achieved, then this would encourage archaeologists to demand and learn these skills.
The final presentation was given by Rachel Opitz (University of Glasgow) on dataARC which has been funded by the National Science Foundation (USA). DataARC is a community backed Research Infrastructure focussed on understanding the interactions between North Atlantic environments and people by linking data and knowledge from thousands of square miles, hundreds of years, and multiple disciplines, from climatology to archaeology, from the humanities to paleoecology. The different partners contribute different data from the SND SEAD database (i.e. pollen and beetles) to the Icelandic Sagas (descriptions of early medieval landscapes) all with the shared aim of:
- enable interdisciplinary synthesis research across the North Atlantic,
- provide access to interdisciplinary datasets that can be easily understood by non-specialists,
- design data discovery, visualization, and integration tools that would encourage responsible use of data from diverse specialisms,
- support research practices that add value to data by making it more usable.
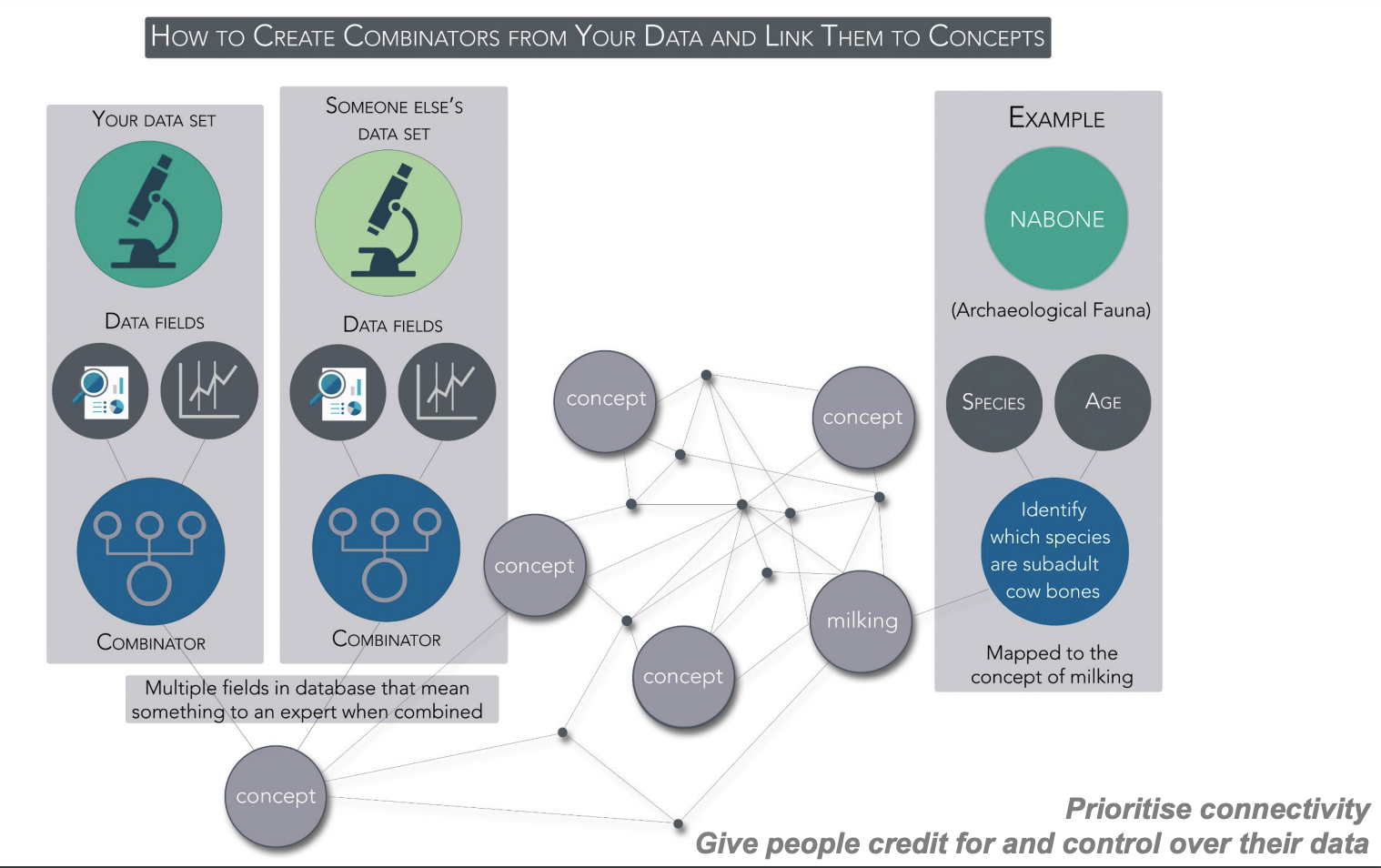
Concept tagging has been used for the Iceland Sagas to help generalists use this information but this has also presented challenges such as coping with ambiguities and messiness, the need for tacit knowledge and wanting the data to be used without it being misused.
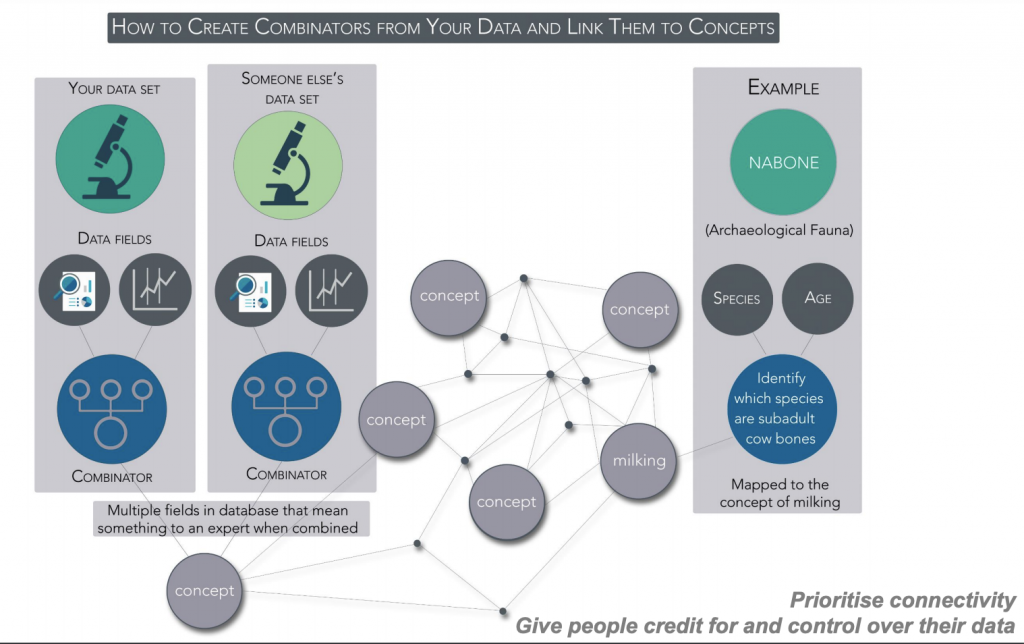
Linking data with combinators mapped to dataARC concepts e.g. “milking”
The approach adopted has been to use combinators which connect datasets using queries for shared concepts. Concept maps are mapped to CIDOC-CRM. Scope notes allow for how different communities interpret the concepts and the result is that multiple roles lead to multiple mappings which have to be shown with explanations. For example, the concept of “driftwood” can be seen as a biological object (related to trees), as a legal object (in relation to resources) and as an actor in an economic system. The interface has been developed to encourage interdisciplinary exploration and meet several goals to:
- emphasise data type (by colour) over specific sources,
- highlight small / long tail data and prevent larger data sources from overwhelming others,
- avoid over interpretation and speculation,
- explain how search results are generated,
- provide motivating examples.
In particular, ARIADNEplus can learn from dataARC how to design research infrastructure such as VREs taking account of various concerns of scholars regarding data sharing and re-use. The key outputs from dataARC in technical terms are the cross-database search tool, Application Programming Interface (API), documentation, extension tools (e.g. ecosystem explorer), and use case examples.
The final discussion ranged over a number of topics and started with a question about how the mapping was done – this is built into the dataARC structure. A question on multiple classification, such as the example of “driftwood”, led to the need of an easy useable tool for annotation in different domain vocabularies. Douglas Tudhope mentioned that such a tool will be developed in ARIADNEplus by the team at University of South-Wales. Doug also suggested that an ARIADNEplus Wikidata type resource could encourage collaboration between research groups employing multiple vocabularies. Moreover, he noted that services are needed to traverse Linked Data of related projects. An example of a case study of data integration was the Dendro Demonstrator in the original ARIADNE project, which focused on data and reports (in different languages) on wooden objects and their dating with dendrochronological techniques.
Planned further workshops
Guntram thanked everyone for attending and announced that two further workshops were planned, the first be on Archaeological Sciences (organised by Sorin Hermon, Cyprus Institute) and the second on Spatial Applications (organised by Benjamin Štular, ZRC-SAZU).